Non-steroidal anti-inflammatory drugs (NSAIDs) are among the most frequently used classes of medications in the world, yet they induce an enteropathy that is associated with high morbidity and mortality. A major limitation to better understanding the pathophysiology and diagnosis of this enteropathy is the difficulty of obtaining information about the primary site of injury, namely the distal small intestine. We investigated the utility of using mRNA from exfoliated cells in stool as a means to surveil the distal small intestine in a murine model of NSAID enteropathy. Specifically, we performed RNA-Seq on exfoliated cells found in feces and compared these data to RNA-Seq from both the small intestinal mucosa and colonic mucosa of healthy control mice or those exhibiting NSAID-induced enteropathy. Global gene expression analysis, data intersection, pathway analysis, and computational approaches including linear discriminant analysis (LDA) and sparse canonical correlation analysis (CCA) were used to assess the inter-relatedness of tissue (invasive) and stool (noninvasive) datasets. These analyses revealed that the exfoliated cell transcriptome closely mirrored the transcriptome of the small intestinal mucosa. Thus, the exfoliome may serve as a non-invasive means of detecting and monitoring NSAID enteropathy (and possibly other gastrointestinal mucosal inflammatory diseases).
Bạn đang xem: Transcriptome là gì
Non-steroidal anti-inflammatory drugs (NSAIDs) are among the most frequently consumed pharmaceuticals worldwide because of their anti-inflammatory, anti-neoplastic, and analgesic effects. Their use can result in an enteropathy that has an alarmingly high rate of morbidity and mortality. In the United States alone, NSAID enteropathy results in approximately 100,000 hospitalizations and 16,500 deaths each year1. An additional 2/3 of both short- and long-term NSAID web4_users develop subclinical or undiagnosed distal small intestinal lesions2. Although detection and management of NSAID-induced lesions of the proximal GI tract (i.e., gastropathy) are well documented, diagnosis and treatment of NSAID-induced damage to the GI tract distal to the duodenum (also known as NSAID enteropathy, affecting primarily the distal jejunum and ileum) remain elusive3,4. This is noteworthy because the incidence of NSAID enteropathy is expected to increase as a result of greater use of NSAIDs to treat rising numbers of inflammatory conditions, to meet the needs of aging populations in North America and Europe, and for their anti-neoplastic effects5. The lower GI tract of multiple mammalian species is affected by NSAIDs in a similar manner in terms of anatomic location, pathological findings, and severity of clinical signs6,7,8.
The pathophysiology of NSAID enteropathy is complex and poorly understood9. Deleterious effects of NSAIDs on the intestinal mucosa including enterocyte cell death, increased mucosal permeability, and interaction of the damaged mucosa with luminal contents including bacteria (i.e., GI microbiota) and bacterial products or components such as lipopolysaccharide (LPS)4,10 has been proposed. The resulting inflammatory cascade is mediated by the innate immune response to LPS and several pro-inflammatory cytokines including tumor necrosis factor (TNF), interleukin (IL)-1, and IL-611,12,13. Although the GI microbiota has recently been implicated as an important contributor to NSAID enteropathy, the precise mechanisms of host-microbiota interactions remain to be elucidated14,15,16,17,18.
An important limitation to understanding the pathogenesis of NSAID enteropathy is the difficulty in obtaining longitudinal (sequential) data from individuals regarding intestinal function and health. A great clinical and investigative need exists to develop non-invasive methods to characterize the health and function of the GI tract distal to the stomach to more effectively identify, study, and manage NSAID enteropathy. A potential strategy to address this limitation is the use of exfoliated intestinal epithelial cells (IECs) and other cell types found in voided stool. Approximately 1/3 of human colonic epithelial cells (up to 1010 cells in an adult) are exfoliated and shed in feces each day19. Isolation and sequencing of the mRNA (host transcriptome) from exfoliated cells has been validated in the context of colon carcinogenesis in rats and humans, and in characterizing human neonatal gastrointestinal development20,21,22,23,24. Exfoliated cells, however, have not been used to evaluate a disease affecting the small intestine. Thus, the objective of this study was to determine whether exfoliated cells could be used as a non-invasive method for detecting and studying NSAID enteropathy in a murine model. Specifically, we performed RNA-Seq on colonic and small intestinal mucosa and exfoliated host cells in feces. We then applied computational approaches, e.g., linear discriminant analysis (LDA) and sparse canonical correlation analysis (CCA), to analyze the inter-relatedness of these data. The goals of these studies were to provide proof-of-principle that the exfoliated cell transcriptome (i.e., the exfoliome) could be used to gain information about NSAID-induced small intestinal injury. Our specific aims were: 1) to determine whether the transcriptome of exfoliated cells reflected gene expression of the small intestinal mucosa; 2) to demonstrate that the transcriptome of exfoliated cells can be used to differentiate healthy and diseased phenotypes; and, 3) to generate hypotheses regarding key biological pathways and processes involved in the pathogenesis of NSAID enteropathy.
The lumen of the GI tract is a formidable environment for RNA transcripts, particularly the small intestine because of the abundance of both host and microbial enzymes and the longer transit time in the SI relative to the colon. Thus, after extracting RNA from tissues and exfoliated cells we examined RNA quality owing to the potential for degradation of mRNA from exfoliated SI cells in stool as it passes through the GI tract. As expected, Bioanalyzer results revealed lower quality RNA in the exfoliated cells than in the tissue (Figure S1a–c). Bioanalyzer traces show that the majority of RNA in the exfoliated cell samples is of microbial origin (23S and 16S rRNA subunits) (Figure S1b). However, due to the oligo dT probe used in the first step of library construction, the mouse transcripts were selectively targeted for cDNA production and subsequent library content. Fastqc results of the subsequent sequenced transcripts revealed high fidelity and quality with all fecal and tissue samples (Figure S2a,b).
Sequencing of these data revealed that the RNA sequencing reads for SI mucosa, colonic mucosa, and exfoliated cells mapped to an average of 19,324 genes, 20,743 genes, and 13,944 genes per sample, respectively (Figure S3a–c). Genes present in low abundance (i.e., ≤4 animals or ≤50 times) across all samples were removed from all datasets and the remaining genes subjected to downstream analysis. This reduced the number of transcripts for downstream analysis in the SI to an average of 17,229 genes, the colon data to 17,244 genes, and the exfoliated data to 10,865 genes per sample. Although filtering reduced the total number of genes in the SI and colon (by 11% and 17%, respectively) less than in the exfoliated cells (22%), the total number of reads was negligibly affected in all datasets (SI reduced from 273,065,200 reads across all samples to 273,026,424 – a 0.001% reduction; colon data reduced from 389,222,037 to 389,122,199 – a 0.003% reduction; exfoliated cell data reduced from 38,292,084 to 38,160,589 – a 0.003% reduction) (Figure S3d–f).
After filtering to remove genes present in low abundance, we examined scatter plots of log(2) counts per million (CPM) for each gene in the 2 treatment groups (i.e., NSAID and control), comparing the exfoliome to the SI and colonic transcriptome in a pairwise manner (Figure S4a,b). There was strong and significant correlation of the CPM data for each of the pairwise comparisons (Spearman’s correlation coefficient; R value > 0.8 and P 1A,B). There was obvious variation between sources (i.e., exfoliated cells, SI, or colonic RNA) in total mammalian reads and distribution of counts. This difference was thought to be due to microbial RNA contamination of the RNA extracted from exfoliated cells (Figure S1b) resulting in fewer reads mapping to the mouse genome. To confirm this, we extracted total counts of 532 genes that have been previously identified as housekeeping genes from each sample and plotted those relative to total gene counts25. Examination of the relative abundance of these genes in each sample revealed that between-sample total count differences were represented by similar magnitudes of differences in abundance of these 532 house-keeping genes, indicating these differences were due to smaller library size attributable to mammalian transcripts and not to sequencing artifact (Fig. 1C).
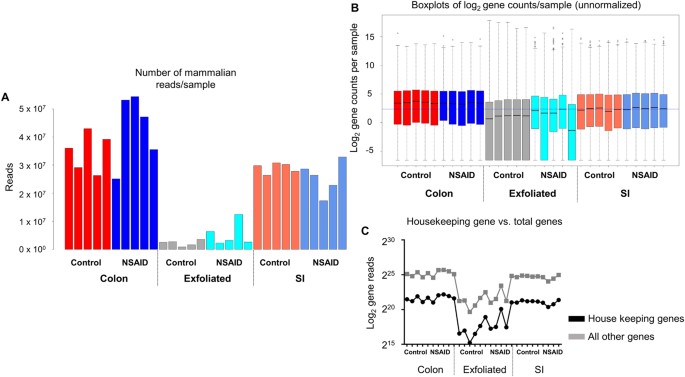
The exfoliome contains fewer reads attributable to the mouse genome than the tissue transcriptomes due to bacterial RNA contamination: (A) Number of mammalian reads per sample for each animal and data source colored by treatment group. (B) Log(2) counts per gene per sample across all treatment groups from the sequenced RNA colored by treatment group. (C) Log(2) total gene reads of 532 murine house-keeping genes (black) and all other genes (grey) per sample across all animals and treatment groups from each data source.
Given the magnitude of differences in library size attributable to mammalian transcripts between the exfoliome and the tissue transciptome, it was necessary to perform the remainder of the analyses separately for each source of RNA (i.e., SI, colon, or exfoliated cells). To account for between-sample variation in read-counts, RNA-Seq data for each dataset were normalized with edgeR accounting for group effects using the function calcNormFactors and the upper-quartile method. Total gene-counts and boxplots of the number of reads/gene for each sample of the normalized data for both tissue and exfoliated cell datasets are shown in Fig. 2A–F. Variability in abundance of post-normalization total house-keeping genes was also improved (Figure S5). Prior to identifying differentially expressed (DE) genes, we assessed biological variability in the exfoliome relative to the tissue transcriptomes. Biological coefficient of variation (BCV) versus the mean log counts per million (CPM) and multi-dimensional scaling (MDS) plots were used to visually assess the similarity of the samples within each treatment group (Fig. 3A–F). These results demonstrate a relatively high degree of variation in the exfoliome (common dispersion = 0.592 and BCV = 0.769) (Fig. 3B) as compared to both tissue transcriptomes (Fig. 3A and C), with a common dispersion of 0.143 and BCV of 0.379 for the SI and common dispersion of 0.126 and BCV of 0.329 for the colon. Notably, MDS based on BCV revealed clear separation of the treatment groups in both the SI transcriptome and exfoliome but not the colonic transcriptome (Fig. 3D–F).
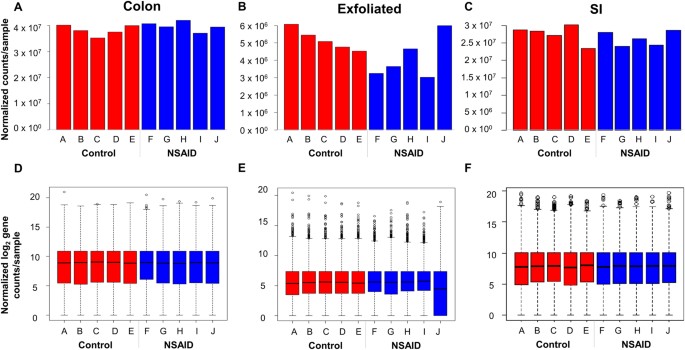
Raw data after filtering and normalization show that the between sample variation in exfoliated cell reads is improved and similar to tissue reads. Total gene counts after normalization for each sample across all treatment groups from the sequenced RNA extracted from (A) colonic mucosa, (B) exfoliated cells and (C) SI mucosa. Normalized log(2) counts per gene per sample across both treatment groups from sequenced RNA extracted from (D) colonic mucosa, (E) exfoliated cells and (F) SI mucosa.
Xem thêm: Là Gì? Nghĩa Của Từ Solvency Là Gì ? Phân Biệt Với Thanh Khoản
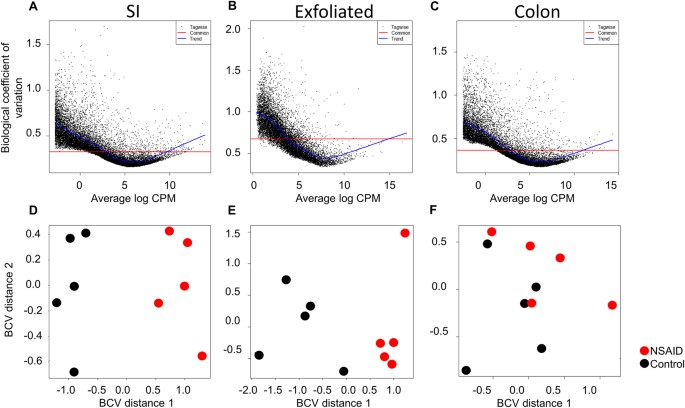
SI transcriptome and exfoliome cluster by treatment group in contrast to colonic transcriptome and biological variability is higher in the exfoliome than the tissue transcriptomes. Biological coefficient of variation (BCV) versus the mean log counts per million (CPM) of the SI transcriptome (A), exfoliome (B) and colonic transcriptome (C). (D) Treatment-based multi-dimensional scaling (MDS) plots of the SI transcriptome and that of the exfoliome (E) and colon (F).
Prior to analyzing these data in order to derive biological meaning, we first wished to determine the anatomic origin of the cellular siggocnhintangphat.com derived from the exfoliome. In order to determine the source of this siggocnhintangphat.com we extracted the counts of genes previously identified and expressed predominantly in specific anatomic locations (i.e. stomach, pancreas, small intestine, and colon). Interestingly, we found that the exfoliome contained virtually no reads from genes representing the stomach or pancreas. In contrast, there was a clear siggocnhintangphat.com arising from both the colon and small intestine (Fig. 4A). As expected genes representing the colon and small intestine were heavily represented in the transcriptomes arising from those locations with some overlap. Similarly, in addition to anatomic origin we also wished to determine the cell types represented in the exfoliome. Clearly, the intestinal mucosa is comprised not only of IECs but also stem cells, crypt cells, goblet cells, Paneth cells (SI), as well as a host of infiltrating immune cells depending on depth of the sample (i.e., lamina propria) and disease state of the GI tract (e.g., inflammation vs. homeostasis). To try to determine the cell types present in these data, we reviewed the literature for marker genes expressed either solely by a specific cell type or at least highly enriched in a specific cell type26,27,28,29,30,31,32,33,34,35,36,37,38. In particular, we extracted the numbers of reads in each sample across all 3 datasets for the following cell types: intestinal stem cells, IECs, crypt base columnar cells, Paneth cells, tuft cells, goblet cells, macrophages, lymphocytes, neutrophils, and smooth muscle. A list of the genes used as biomarkers for each of these cell types is shown in Table S1. Interestingly, we found that all cell types were present in all datasets as identified by the presence of at least 2 marker genes per cell type (Fig. 4B). Visually there were minimal differences among the 3 datasets with the expected exception of fewer reads in the exfoliated cell data and absence of a few marker genes of intestinal stems cells in the exfoliome with concurrent low expression in the tissue transcriptomes. These data suggest that the mucosal transcriptome and exfoliome represent siggocnhintangphat.coms from similar cell types that comprise not only IECs but reads from the diverse array of cell types expected to be found in the intestinal mucosa.
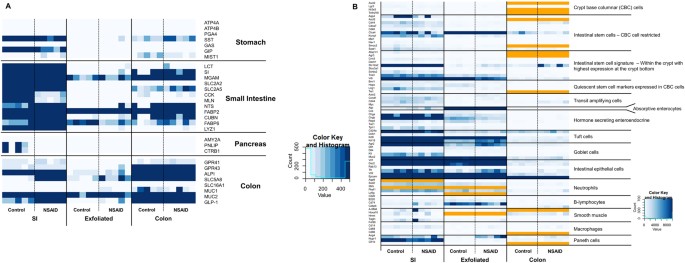
(A) The exfoliome siggocnhintangphat.com arises from cells sloughed from both the small intestine and colon and comprises reads from the diverse array of cell types expected to be found in the intestinal mucosa (A) Heatmap showing counts of genes that are reported to be primarily expressed at specific anatomic locations (stomach, pancreas, small intestine, colon). All genes with counts greater than 400 are colored dark blue. (B) Heatmap showing counts of biomarker genes from each sample and each data source (orange = gene not expressed).
After characterizing the cellular source of the siggocnhintangphat.coms derived from these datasets, we examined each dataset for alterations induced by NSAIDs by comparing the transcriptome (or exfoliome) between the control group and NSAID group. In human subjects and preclinical models, NSAID-induced lower GI damage primarily occurs in the distal jejunum and ileum3,4. As expected, we observed marked pathological findings in the distal SI but no notable microscopic abnormalities in the colon in NSAID-treated mice (Fig. 5A). Therefore, RNA extracted from SI mucosal scrapings of NSAID-treated subjects should demonstrate marked mucosal pathology whereas the RNA from colonic mucosal scrapings should reflect minimal pathology.
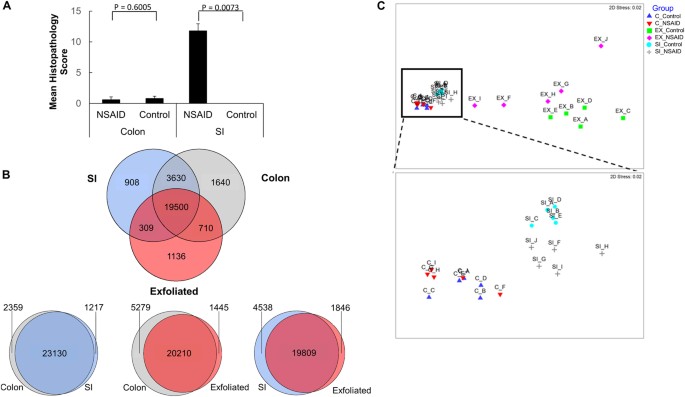
Microscopic pathology reveals NSAID injury is confined to the SI. Despite great overlap between the three RNA-Seq datasets, the exfoliome distinguishes between NSAID and control animals similar to the SI transcriptome, whereas the colonic transcriptome does not. (A) Microscopic pathology scores from colon and small intestinal mucosa in control mice and NSAID-treated mice. (B) Venn diagram showing intersection of gene lists among the datasets. (C) Multi-dimensional scaling plot of each sample color-coded by source and treatment group. Inset of panel C enlarged to show degree of separation of groups in the SI transcriptome and lack of clear separation in colonic transcriptome.
In order to determine whether the exfoliome more closely resembled the colonic transcriptome or the SI transcriptome, we first crudely examined the intersection of gene lists from each source. This analysis revealed >90% overlap among the 3 datasets in terms of presence and absence of genes (Fig. 5C). Despite this overlap of genes, non-metric multidimensional scaling (NMDS) plots demonstrated differences among these 3 datasets (Fig. 5C and inset). Although the exfoliated cell data differed from the SI and colon data because of smaller library size and fewer mammalian reads potentially resulting from degradation of RNA in the GI tract, evidence of clustering of the treatment groups was observed in the SI and exfoliated cell data but was absent in the colonic data (Fig. 5C and inset). Analysis of similarity (ANOSIM) based on the Bray-Curtis dissimilarity metric quantitatively demonstrated differences between NSAID and control groups for the SI (R value = 0.760; P = 0.008) and exfoliated cells (R value 0.280; P = 0.024) but not for the colonic data (R value 0.194; P = 0.090).
To further examine the interdependent relationship between these 3 transcriptomic profiles, we utilized sparse CCA, a novel multivariate statistical analysis approach. Sparse CCA is a dimensionality reduction technique that identifies the fewest numbers of genes that show the greatest amount of correlation between datasets according to specific optimality criteria. Although the sparse CCA plots should not be assigned any particular biological interpretation, they can be considered a stringent method for determining correlation of large datasets39. As revealed by NMDS plots, sparse CCA plots demonstrated that the transcriptome from exfoliated cells correlated well with the SI transcriptome in these mice, and that the SI and exfoliated cell datasets discriminated NSAID-treated from control groups, whereas the colonic mucosal transcriptome did not (Fig. 6A–D).
Sparse canonical correlation analysis (CCA) reveals that the global transcriptome profiles from exfoliated cells correlates well with the transcriptome profile of the SI. In contrast, the colonic transcriptome data do not discriminate well between treatment groups. Sparse CCA plots positioned by 1st and 2nd component scores from (exfoliated cells and colored by the 1st component SI scores, (B) SI and colored by 1st component scores from exfoliated cells, (C) exfoliated cells and colored by the 1st component colon scores and (D) colonic mucosa and colored by the 1st component exfoliated scores.
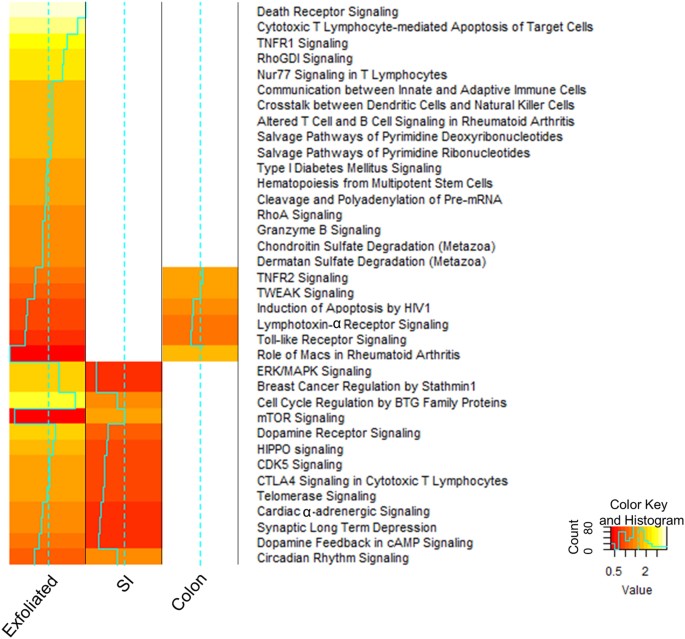
We next examined the similarities and differences in the gene expression profiles from exfoliated cells compared with those from the scraped intestinal mucosa. We identified DE genes between control and NSAID-treated mice for each dataset. Interestingly, both the exfoliome and SI transcriptome had >1000 DE genes (FDR P 7A). Venn diagrams revealed sparse overlap (12%) of the DE expressed genes between the SI transcriptome and the exfoliome and even less overlap between the colonic transcriptome and the exfoliome (6%; Fig. 7A). Despite this sparse overlap, the pathways enriched in the exfoliome and SI transcriptome were similar whereas there was much less similarity between the exfoliome and colonic transcriptome. Specifically, IPA Ingenuity Knowledgebase (www.ingenuity.com) pathway analysis revealed both SI and exfoliated cell datasets exhibited similar occupancy and predicted directionality (Z-score) in the canonical pathways represented (Fig. 7B). In contrast, few pathways were represented in the colonic data (Fig. 7B). For example, Toll-like receptor signaling (which is known to play a crucial role in the pathogenesis of NSAID enteropathy)9,10,13,18,40,41 was upregulated by NSAID administration in both the exfoliome and small intestinal transcriptome but genes related to this pathway were not altered in the colonic transcriptome resulting in no occupancy of this canonical pathway. Indeed, the proportion of pathways represented in the colon (31%; 21/67) was significantly less than that of the exfoliome (84%; 56/67; P S6). Specifically, there were significantly fewer upstream regulators expressed in the colon (41%; 780/1888) than in either the SI (73%; 1380/1888; P 8A–E)42. These plots confirmed lack of overlap between DE genes within each dataset and show that the DE genes within the exfoliome had a greater effect-size than those within the tissue.
Xem thêm: Những Bài Hát Do Ca Sĩ Various Artists Là Gì Của Nhau? 情是什么(蔡琴)/ Tình Là Gì
The exfoliated cell transcriptome is similar to the tissue transcriptome as shown by overlapping gene lists and pathways. (A) Venn diagram of the intersection of differentially expressed genes found in the exfoliome and tissue transcriptomes. (B) Heat map of the Z-scores of the canonical pathways to which the differentially expressed genes between control and NSAID-treated animals were mapped from the SI transcriptome (left column), exfoliome (middle column) and the colonic transcriptome (right column).
MA plots demonstrate the expression of genes identified as differentially expressed (DE) in the exfoliome and in the tissue transcriptomes. MA plot of genes from the (A) SI transcriptome, (B) exfoliome and (C) colon transcriptome with DE genes (FDR Full size image
$$({u}_{1},{v}_{1})=mathop{{rm{argmax}}}limits_{u,v},{rm{Corr}}({u}^{T}x,{v}^{T}y)=mathop{{rm{argmax}}}limits_{u,v}frac{{u}^{T}{{rm{Sigma }}}_{xy}nu }{sqrt{({u}^{T}{{rm{Sigma }}}_{xx}nu )({v}^{T}{{rm{Sigma }}}_{yy}nu )}},$$
Chuyên mục: Định Nghĩa







